Kernel Interpolation with RKeOps
Amelie Vernay
Chloe Serre-Combe
Ghislain Durif
2024-02-13
Source:ci/vignettes/Kernel_Interpolation_rkeops.Rmd
Kernel_Interpolation_rkeops.RmdSetup
library(ggplot2) # graph generation
library(dplyr) # data.frame manipulation
library(pracma) # to create the meshgrid
library(plotly) # reactive graph generation
library(reshape) # matrix to data.frame reformatting
# load rkeops
library(rkeops)
# create a dedicated Python environment with reticulate (to be done only once)
reticulate::virtualenv_create("rkeops")
# activate the dedicated Python environment
reticulate::use_virtualenv(virtualenv = "rkeops", required = TRUE)
# install rkeops requirements (to be done only once)
install_rkeops()
# use 64bit floating operation precision
rkeops_use_float64()
# check rkeops install
check_rkeops()
## 'keopscore' is available.
## 'pykeops' is available.
## pyKeOps with numpy bindings is working!
## 'rkeops' is ready and working.
## [1] TRUE
# use 64bit floating operation precision
rkeops_use_float64()For build on CRAN, use CPU computing with 2 cores:
rkeops_use_cpu(ncore = 2)Note: you can run rkeops_use_cpu() to
use all available CPU cores or rkeops_use_gpu() to enable
GPU computing.
Introduction
This tutorial is highly inspired by the PyKeOps tutorial on kernel interpolation.
The goal here is to solve a linear system of the form,
\[ \begin{aligned} &a^{*} = \underset{a}{\textrm{argmin}} \frac{1}{2} \langle a, (\lambda \textrm{Id} + K_{xx})a \rangle - \langle a, b \rangle \\ \text{i.e.} \quad &a^{*} = (\lambda \textrm{Id} + K_{xx})^{-1}b \end{aligned} \]
where \(K_{xx}\) is a symmetric,
positive definite matrix encoded as an RKeOps LazyTensor,
and \(\lambda\) is a nonnegative
regularization parameter. In the following script, we use the conjugate
gradient method to solve large-scale Kriging (a.k.a. Gaussian process
regression or generelized
spline interpolation) problems with a linear memory
footprint.
Interpolation in 1D
Generate some data:
N <- 10000 # number of samples
x <- matrix(runif(N * 1), N, 1)
pert <- matrix(runif(N * 1), N, 1) # random perturbation to create b
# Some random-ish 1D signal:
b <- x + 0.5 * sin(6 * x) + 0.1 * sin(20 * x) + 0.05 * pertSpecify our regression model - a simple
Gaussian variogram or kernel matrix of
deviation sigma.
gaussian_kernel <- function(x, y, sigma = 0.1) {
x_i <- Vi(x) # symbolic 'i'-indexed matrix
y_j <- Vj(y) # symbolic 'j'-indexed matrix
D_ij <- sum((x_i - y_j)^2) # symbolic matrix of squared distances
res <- exp(-D_ij / (2 * sigma^2)) # symbolic Gaussian kernel matrix
return(res)
}Kernel Interpolation
We implement the conjugate gradient algorithm, that includes the
lambda regularization parameter.
CG_solve <- function(K, b, lambda, eps = 1e-6) {
# ----------------------------------------------------------------
# Conjugate gradient algorithm to solve linear systems of the form
# (K + lambda * Id) * a = b.
#
# K: a LazyTensor encoding a symmetric positive definite matrix
# (the spectrum of the matrix must not contain zero)
# b: a vector corresponding to the second member of the equation
# lambda: Non-negative ridge regularization parameter
# (lambda = 0 means no regularization)
# eps (default=1e-6): precision parameter
# ----------------------------------------------------------------
delta <- length(b) * eps^2
a <- 0
r <- b
nr2 <- sum(r^2) # t(r)*r (L2-norm)
if(nr2 < delta) {
return(0 * r)
}
p <- r
k <- 0
while (TRUE) {
Mp <- K %*% Vj(p) + lambda * p
alp <- nr2 / sum(p * Mp)
a <- a + (alp * p)
r <- r - (alp * Mp)
nr2new <- sum(r^2)
if (nr2new < delta) {
break
}
p <- r + (nr2new / nr2) * p
nr2 <- nr2new
k <- k + 1
}
return(a) # should be such that K%*%a + lambda * Id * a = b (eps close)
}Perform the Kernel interpolation, without forgetting
to specify the ridge regularization parameter lambda which
controls the trade-off between a perfect fit (lambda =
\(0\)) and a smooth interpolation
(lambda = \(+\infty\)):
K_xx <- gaussian_kernel(x, x)
lambda <- 1
start <- Sys.time()
a <- CG_solve(K_xx, b, lambda = lambda)
## Loading required namespace: testthat
end <- Sys.time()
time <- round(as.numeric(end - start), 5)
print(paste("Time to perform an RBF interpolation with",
N,"samples in 1D:", time, "s.",
sep = " "
)
)
## [1] "Time to perform an RBF interpolation with 10000 samples in 1D: 21.58736 s."Display the (fitted) model on the unit interval:
# extrapolate on a uniform sample
t <- as.matrix(seq(from = 0, to = 1, length.out = N))
K_tx <- gaussian_kernel(t, x)
mean_t <- K_tx %*% Vj(a)
D <- as.data.frame(cbind(x, b, t, mean_t))
colnames(D) <- c("x", "b", "t", "mean_t")
# 1D plot
ggplot(aes(x = x, y = b), data = D) +
geom_point(color = '#1193a8', alpha = 0.5, size = 0.4) +
geom_line(aes(x = t, y = mean_t), color = 'darkred') +
annotate("text", x = .75, y = .1,
label = paste("Number of samples: ", N,
sep = "")
) +
theme_bw()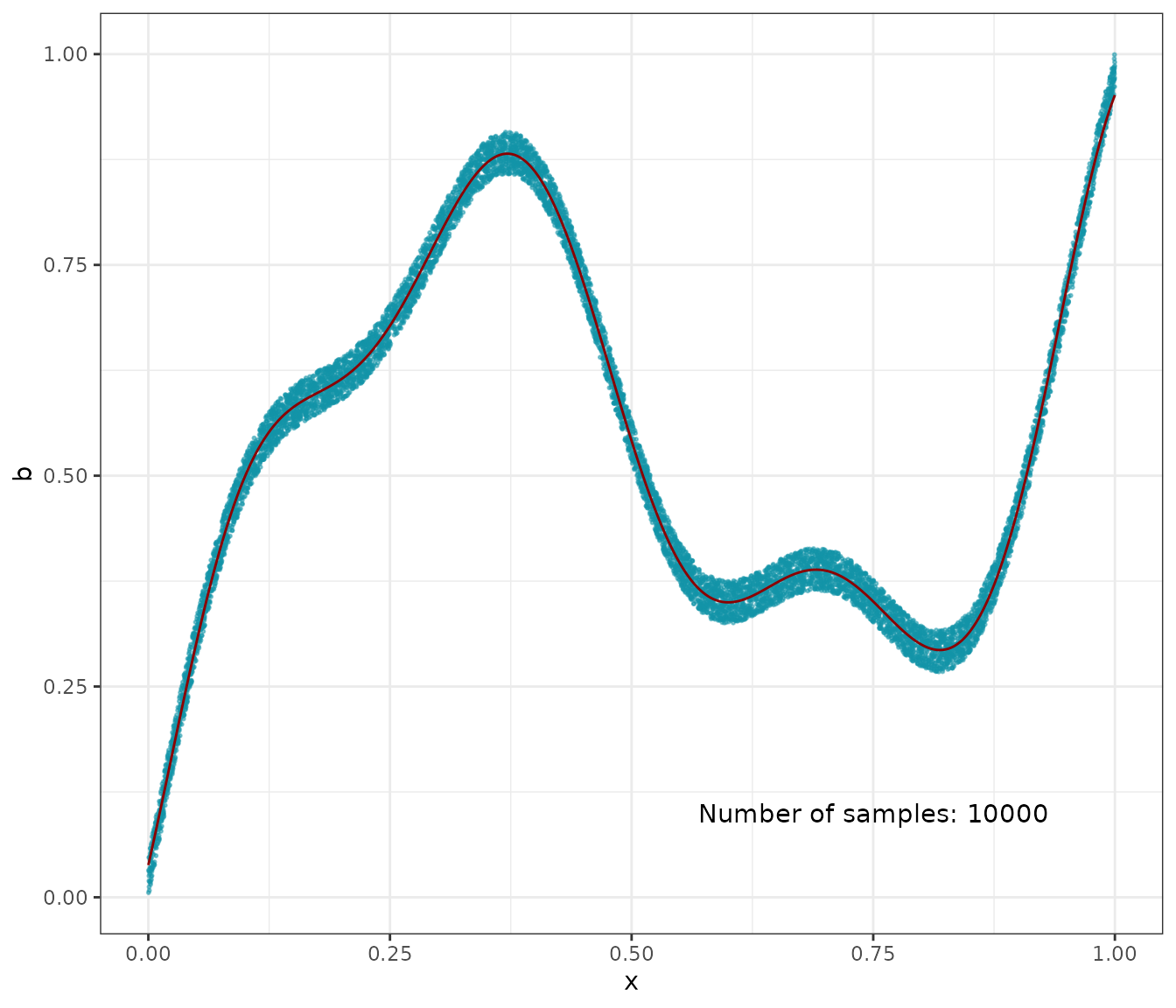
Interpolation in 2D
Generate some data:
N <- 10000
# Sampling locations:
x <- matrix(runif(N * 2), N, 2)
# Some random-ish 2D signal:
b <- as.matrix(rowSums((x - 0.5)^2))
b[b > 0.4^2] = 0
b[b < 0.3^2] = 0
b[b >= 0.3^2] = 1
pert <- matrix(runif(N * 1), N, 1) # random perturbation to create b
b <- b + 0.05 * pert
# Add 25% of outliers:
Nout <- N %/% 4
b[(length(b) - Nout + 1):length(b)] <- matrix(runif(Nout * 1), Nout, 1)Specify our regression model - a simple
Exponential variogram or Laplacian
kernel matrix of deviation sigma:
laplacian_kernel <- function(x, y, sigma = 0.1) {
x_i <- Vi(x)
y_j <- Vj(y)
D_ij <- sum((x_i - y_j)^2)
res <- exp(-sqrt(D_ij) / sigma)
return(res)
}Perform the Kernel interpolation, without forgetting
to specify the ridge regularization parameter lambda which
controls the trade-off between a perfect fit (lambda =
\(0\)) and a smooth interpolation
(lambda = \(+\infty\)):
lambda <- 10 # Ridge regularization
start <- Sys.time()
K_xx <- laplacian_kernel(x, x)
a <- CG_solve(K_xx, b, lambda = lambda)
end <- Sys.time()
time <- round(as.numeric(end - start), 5)
print(paste("Time to perform an RBF interpolation with",
N, "samples in 2D:", time, "s.",
sep = " "))
## [1] "Time to perform an RBF interpolation with 10000 samples in 2D: 32.48329 s."Interpolate and display the (fitted) model:
# Interpolate on a uniform sample:
X <- seq(from = 0, to = 1, length.out = 100)
Y <- seq(from = 0, to = 1, length.out = 100)
G <- meshgrid(X, Y)
t <- cbind(as.vector(G$X), as.vector(G$Y))
K_tx <- laplacian_kernel(t, x)
mean_t <- K_tx %*% Vj(a)
mean_t <- matrix(mean_t, 100, 100)
mean_t <- mean_t[nrow(mean_t):1, ]
# Data
data2plot_sample <- data.frame(
X = x[,1],
Y = x[,2],
value = b
)
ggplot(data2plot_sample, aes(x = X, y = Y, col = value)) +
geom_point() +
scale_color_gradient(low = "#C2C2C9", high = "darkred") +
coord_fixed() +
theme_bw()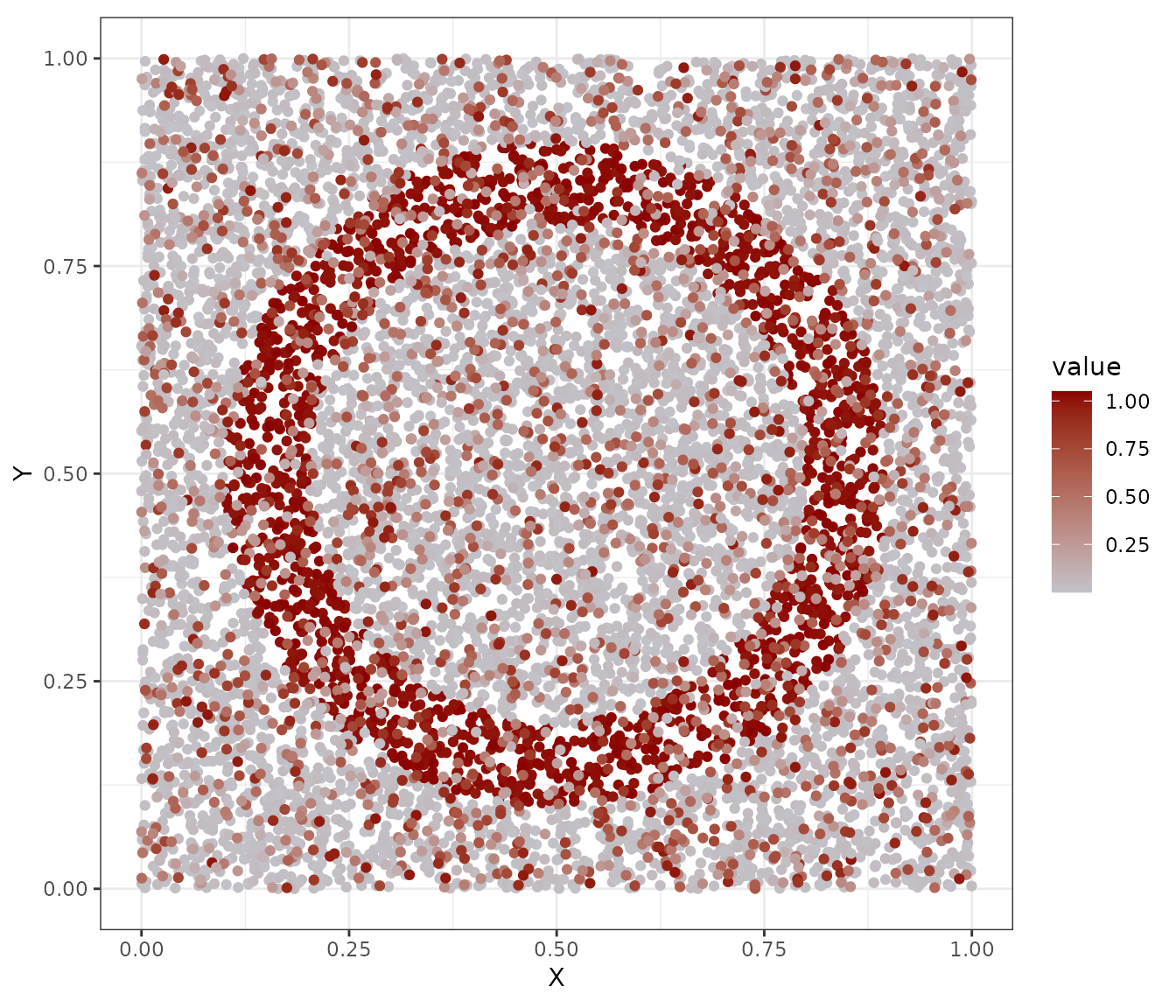
Data sample
# Interpolation
data2plot_interpol <- reshape::melt(mean_t) %>%
mutate(X = X[X1], Y = Y[X2]) %>% dplyr::select(!c(X1, X2))
ggplot(data2plot_interpol, aes(x = X, y = Y, fill = value)) +
geom_tile() +
scale_fill_gradient(low = "#C2C2C9", high = "darkred") +
coord_fixed() +
theme_bw()
Kernel interpolation
Plotly interactive graph:
# 2D plot: noisy samples and interpolation in the background
fig <- plot_ly(z = mean_t,
type = "heatmap",
colors = colorRamp(c("#C2C2C9", "darkred")),
zsmooth ="best"
)
fig <- fig %>% add_trace(type = "scatter",
x = ~(100 * x[, 1]),
y = ~(100 * x[, 2]),
mode = "markers",
marker = list(size = 4, color = as.vector(b))
)
fig <- fig %>% plotly::layout(xaxis = list(title = ""),
yaxis = list(title = ""))
colorbar(fig, limits = c(0, 1), x = 1, y = 0.75)