Note
Go to the end to download the full example code
K-means clustering - NumPy API
The pykeops.numpy.LazyTensor.argmin() reduction supported by KeOps pykeops.numpy.LazyTensor allows us
to perform bruteforce nearest neighbor search with four lines of code.
It can thus be used to implement a large-scale
K-means clustering,
without memory overflows.
Note
For large and high dimensional datasets, this script is outperformed by its PyTorch counterpart which avoids transfers between CPU (host) and GPU (device) memories.
Setup
Standard imports:
import time
import numpy as np
from matplotlib import pyplot as plt
from pykeops.numpy import LazyTensor
import pykeops.config
dtype = "float32" # May be 'float32' or 'float64'
Simple implementation of the K-means algorithm:
def KMeans(x, K=10, Niter=10, verbose=True):
N, D = x.shape # Number of samples, dimension of the ambient space
# K-means loop:
# - x is the point cloud,
# - cl is the vector of class labels
# - c is the cloud of cluster centroids
start = time.time()
c = np.copy(x[:K, :]) # Simplistic random initialization
x_i = LazyTensor(x[:, None, :]) # (Npoints, 1, D)
for i in range(Niter):
c_j = LazyTensor(c[None, :, :]) # (1, Nclusters, D)
D_ij = ((x_i - c_j) ** 2).sum(
-1
) # (Npoints, Nclusters) symbolic matrix of squared distances
cl = D_ij.argmin(axis=1).astype(int).reshape(N) # Points -> Nearest cluster
Ncl = np.bincount(cl).astype(dtype) # Class weights
for d in range(D): # Compute the cluster centroids with np.bincount:
c[:, d] = np.bincount(cl, weights=x[:, d]) / Ncl
end = time.time()
if verbose:
print(
"K-means example with {:,} points in dimension {:,}, K = {:,}:".format(
N, D, K
)
)
print(
"Timing for {} iterations: {:.5f}s = {} x {:.5f}s\n".format(
Niter, end - start, Niter, (end - start) / Niter
)
)
return cl, c
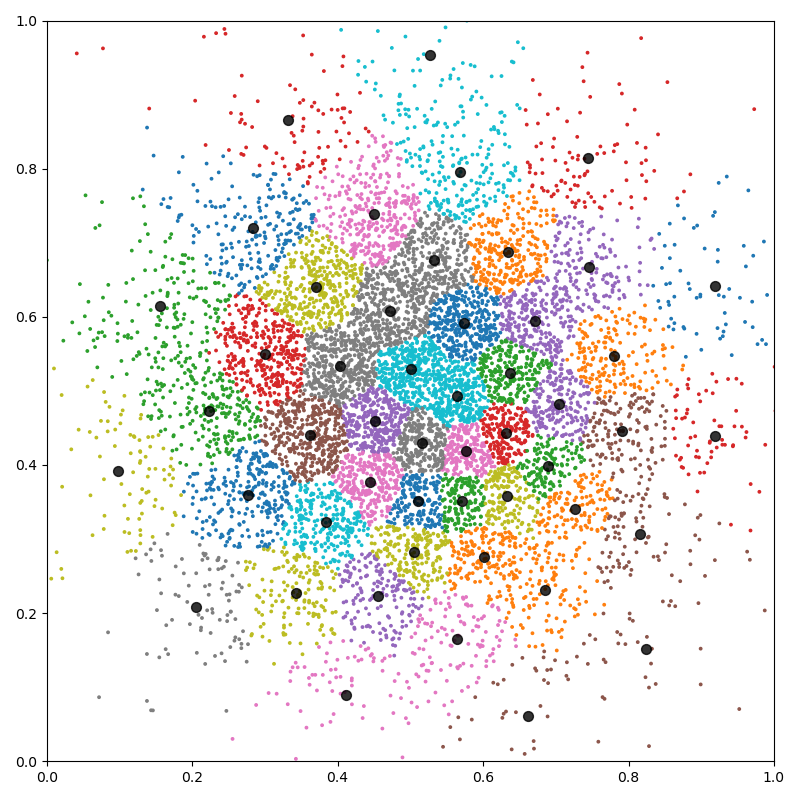
K-means in 2D
First experiment with N=10,000 points in dimension D=2, with K=50 classes:
N, D, K = 10000, 2, 50
Define our dataset:
x = np.random.randn(N, D).astype(dtype) / 6 + 0.5
Perform the computation:
cl, c = KMeans(x, K)
K-means example with 10,000 points in dimension 2, K = 50:
Timing for 10 iterations: 0.00325s = 10 x 0.00033s
Fancy display:
plt.figure(figsize=(8, 8))
plt.scatter(x[:, 0], x[:, 1], c=cl, s=30000 / len(x), cmap="tab10")
plt.scatter(c[:, 0], c[:, 1], c="black", s=50, alpha=0.8)
plt.axis([0, 1, 0, 1])
plt.tight_layout()
plt.show()
K-means in dimension 100
Second experiment with N=1,000,000 points in dimension D=100, with K=1,000 classes:
if pykeops.config.gpu_available:
N, D, K = 1000000, 100, 1000
x = np.random.randn(N, D).astype(dtype)
cl, c = KMeans(x, K)
K-means example with 1,000,000 points in dimension 100, K = 1,000:
Timing for 10 iterations: 9.96571s = 10 x 0.99657s
Total running time of the script: (0 minutes 11.933 seconds)